publications
2026
-
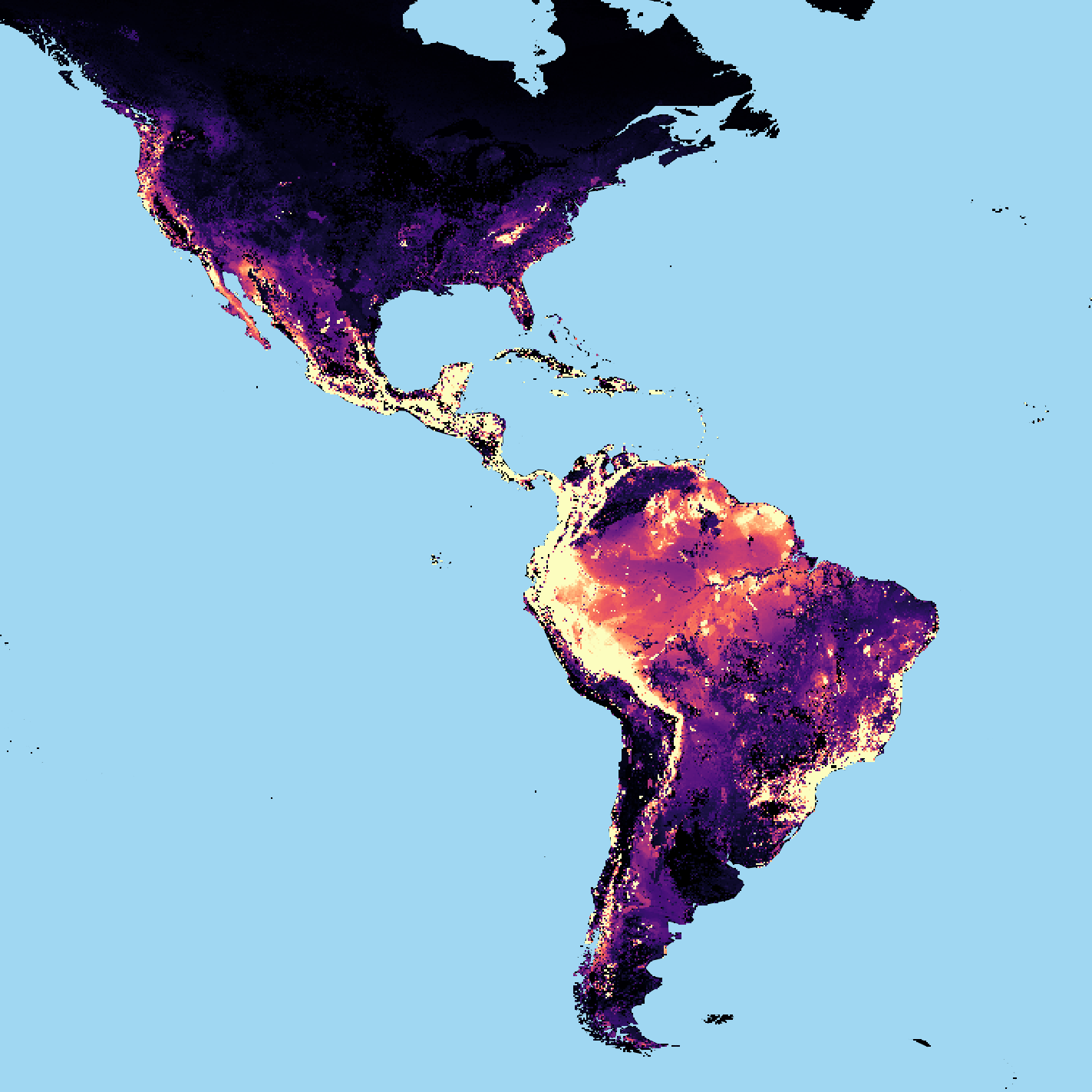 Informing conservation problems and actions using an indicator of extinction risk: A detailed assessment of applying the LIFE metricAlison Eyres , Andy Arnell , Thomas S Ball , and 10 more authorsBiological Conservation, Jan 2026
Informing conservation problems and actions using an indicator of extinction risk: A detailed assessment of applying the LIFE metricAlison Eyres , Andy Arnell , Thomas S Ball , and 10 more authorsBiological Conservation, Jan 2026Extinction is a critical issue, with land-use change the main threat to many terrestrial species. Understanding and tackling this requires global, comparable, and scalable metrics that link land-cover change to extinction risk and are useable across diverse conservation contexts. Here, we demonstrate the flexibility of the new Land-cover change Impacts on Future Extinctions (LIFE) metric through five distinct case studies. First, we explore the near real-time quantification of biodiversity harms in tropical hotspots by integrating LIFE with forest loss data. Second, we couple LIFE with crop distribution and trade data to assess variation in extinction impacts mediated by food consumption – specifically of apples in the UK. Third, we test LIFE’s suitability for use in biodiversity compensation through a hypothetical scenario in Sumatra. Fourth, we use LIFE to prioritize competing conservation investments by comparing benefits of area-based projects in Honduras. Finally, we combine LIFE with counterfactual methods to evaluate the effectiveness of a long-term conservation project in Sierra Leone. Together, these examples show that LIFE offers actionable insights into a geographically and thematically wide range of conservation challenges, from land-use planning to sustainable consumption. Like all global metrics, LIFE’s broad applicability relies on assumptions and simplifications. It should be used cautiously, and alongside local knowledge and ground-truthing, especially for restoration, offsetting, or fine-scale analysis, and in poorly studied areas. By providing an accompanying “How-to” guide, we aim to ensure LIFE can be used widely to inform understanding of the extinction crisis and support tangible actions to halt it.
@article{eyres2026informing, title = {Informing conservation problems and actions using an indicator of extinction risk: A detailed assessment of applying the LIFE metric}, author = {Eyres, Alison and Arnell, Andy and Ball, Thomas S and Cuthbert, Richard J and Dales, Michael and Guizar-Couti{\~n}o, Alejandro and Holland, Jody and Luz-Ricca, Emilio and Madhavapeddy, Anil and Pain, Leila and Swinfield, Tom and White, Thomas B and Balmford, Andrew}, journal = {Biological Conservation}, volume = {315}, pages = {111663}, month = jan, year = {2026}, publisher = {Elsevier}, }
2024
-
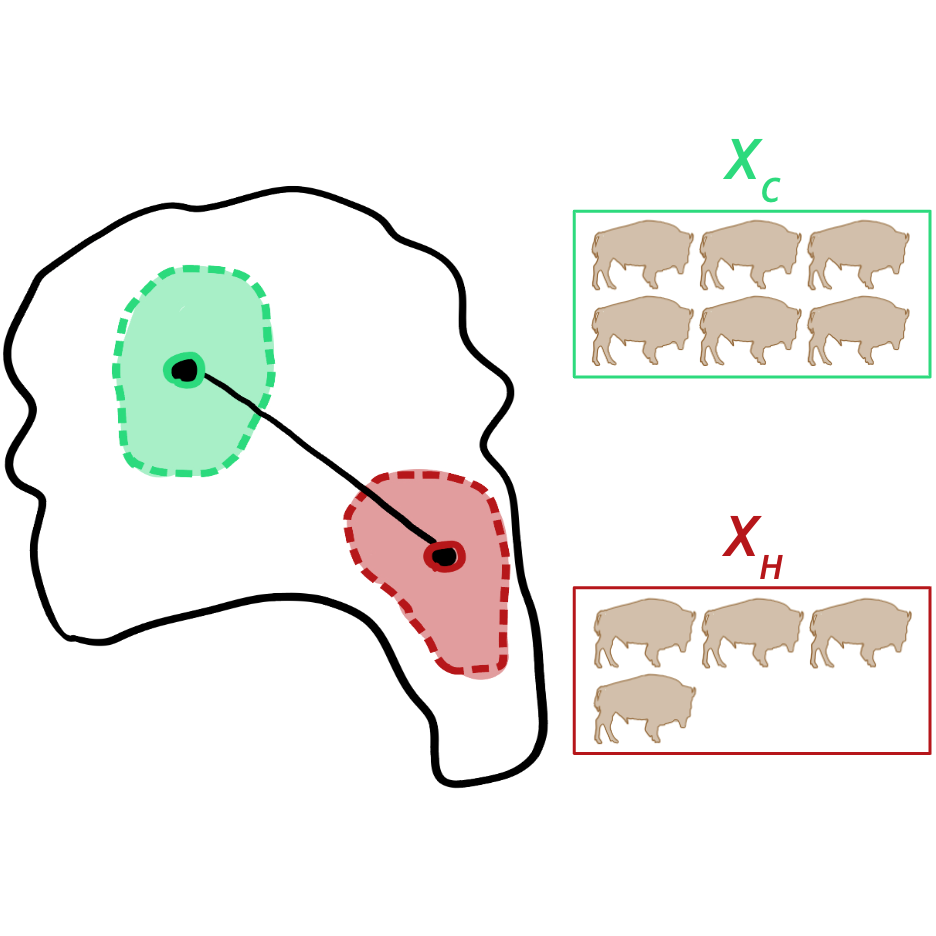 Quantifying species-specific responses to hunting pressureEmilio Luz-RiccaJul 2024
Quantifying species-specific responses to hunting pressureEmilio Luz-RiccaJul 2024We are currently experiencing an unprecedented loss of terrestrial biodiversity, particularly in the species-rich tropics. Species extinctions are primarily driven by loss of habitat, which is relatively easy to monitor by satellite remote sensing; other anthropogenic threats to biodiversity, like hunting, are much more difficult to observe directly. Further, little is known about how the local abundance of animal populations responds to hunting pressure. Recent studies have applied predictive modelling using statistical methods to implicitly assess hunting pressure through collections of tropical mammal and bird abundance responses. However, few predictive models have been considered to date and a thorough assessment of model generalisability is needed. Building on these studies, I present a comprehensive assessment of approaches for this predictive task. In particular, I reproduced the previous state-of-the art (a mixed-effects generalised linear hurdle model), thoroughly tested (nonlinear) predictive methods through application of automated machine learning, experimented with embeddings from pre-trained deep learning models as a supplement to hand-chosen predictors, and closely inspected spatial and taxonomic generalisability using cross-validation. I found that nonlinear hurdle models tend to outperform the existing mixed-effects linear hurdle model baseline, especially when random effects are excluded during prediction. Deep learning embeddings were largely unhelpful as supplemental predictors, but could be used to reliably predict hunting pressure on their own if used with the nonlinear hurdle model. Finally, spatial and taxonomic generalisation remained very difficult for all models tested, but improved in the presence of more training data. Through this work, I advance the state-of-the-art for predicting species-specific abundance responses to hunting pressure in the tropics and provide well-documented, reproducible code to support further predictive benchmarking for this task.
@phdthesis{luzricca2023quantifying, author = {Luz-Ricca, Emilio}, type = {Master's thesis}, title = {Quantifying species-specific responses to hunting pressure}, school = {University of Cambridge}, year = {2024}, month = jul, }
2023
-
 Data-free one-shot federated learning under very high statistical heterogeneityClare Elizabeth Heinbaugh , Emilio Luz-Ricca, and Huajie ShaoIn The Eleventh International Conference on Learning Representations , Feb 2023
Data-free one-shot federated learning under very high statistical heterogeneityClare Elizabeth Heinbaugh , Emilio Luz-Ricca, and Huajie ShaoIn The Eleventh International Conference on Learning Representations , Feb 2023Federated learning (FL) is an emerging distributed learning framework that collaboratively trains a shared model without transferring the local clients’ data to a centralized server. Motivated by concerns stemming from extended communication and potential attacks, one-shot FL limits communication to a single round while attempting to retain performance. However, one-shot FL methods often degrade under high statistical heterogeneity, fail to promote pipeline security, or require an auxiliary public dataset. To address these limitations, we propose two novel data-free one-shot FL methods: FedCVAE-Ens and its extension FedCVAE-KD. Both approaches reframe the local learning task using a conditional variational autoencoder (CVAE) to address high statistical heterogeneity. Furthermore, FedCVAE-KD leverages knowledge distillation to compress the ensemble of client decoders into a single decoder. We propose a method that shifts the center of the CVAE prior distribution and experimentally demonstrate that this promotes security, and show how either method can incorporate heterogeneous local models. We confirm the efficacy of the proposed methods over baselines under high statistical heterogeneity using multiple benchmark datasets. In particular, at the highest levels of statistical heterogeneity, both FedCVAE-Ens and FedCVAE-KD typically more than double the accuracy of the baselines.
@inproceedings{heinbaugh2023data, title = {Data-free one-shot federated learning under very high statistical heterogeneity}, author = {Heinbaugh, Clare Elizabeth and Luz-Ricca, Emilio and Shao, Huajie}, booktitle = {The Eleventh International Conference on Learning Representations}, year = {2023}, month = feb, } -
 Automating sandhill crane counts from nocturnal thermal aerial imagery using deep learningEmilio Luz-Ricca, Kyle Landolt , Bradley A Pickens , and 1 more authorRemote Sensing in Ecology and Conservation, Apr 2023
Automating sandhill crane counts from nocturnal thermal aerial imagery using deep learningEmilio Luz-Ricca, Kyle Landolt , Bradley A Pickens , and 1 more authorRemote Sensing in Ecology and Conservation, Apr 2023Population monitoring is essential to management and conservation efforts for migratory birds, but traditional low‐altitude aerial surveys with human observers are plagued by individual observer bias and risk to flight crews. Aerial surveys that use remote sensing can reduce bias and risk, but manual counting of wildlife in imagery is laborious and may be cost‐prohibitive. Therefore, automated methods for counting are critical to cost‐efficient application of remote sensing for wildlife surveys covering large areas. We conducted nocturnal surveys of sandhill cranes (Antigone canadensis) during spring migration in the Central Platte River Valley of Nebraska, USA, using midwave thermal infrared sensors. We developed a framework for automated counting of sandhill cranes from thermal imagery using deep learning, assessed and compared the performance of two automated counting models, and quantified the effect of spatial resolution on counting accuracy. Aerial thermal imagery data were collected in March 2018 and 2021; 40 images were analyzed. We applied two deep learning models: an object detection approach, Faster R‐CNN and a recently developed pixel‐density estimation approach, ASPDNet. Model performance was determined using data independent of the training imagery. The effect of spatial resolution was quantified with a beta regression on relative error. Our results showed model accuracy of 9% mean percent error for ASPDNet and 1\8% for Faster R‐CNN. Most error was related to the undercounting of sandhill cranes. ASPDNet had <50% of the error of Faster R‐CNN as measured by mean percent error, root‐mean‐squared error and mean absolute error. Spatial resolution affected accuracy of both models, with error rate increasing with coarser resolution, particularly with Faster R‐CNN. Deep learning models, particularly pixel‐density estimators, can accurately automate counting of migratory birds in a dense, aggregate setting such as nocturnal roosting sites.
@article{luz2023automating, title = {Automating sandhill crane counts from nocturnal thermal aerial imagery using deep learning}, author = {Luz-Ricca, Emilio and Landolt, Kyle and Pickens, Bradley A and Koneff, Mark}, journal = {Remote Sensing in Ecology and Conservation}, volume = {9}, number = {2}, pages = {182--194}, year = {2023}, month = apr, publisher = {Wiley Online Library}, }
2022
-
 Blinking-based multiplexing: a new approach for differentiating spectrally overlapped emittersGrace A DeSalvo , Grayson R Hoy , Isabelle M Kogan , and 5 more authorsThe Journal of Physical Chemistry Letters, Jun 2022
Blinking-based multiplexing: a new approach for differentiating spectrally overlapped emittersGrace A DeSalvo , Grayson R Hoy , Isabelle M Kogan , and 5 more authorsThe Journal of Physical Chemistry Letters, Jun 2022Multicolor single-molecule imaging is widely applied to answer questions in biology and materials science. However, most studies rely on spectrally distinct fluorescent probes or time-intensive sequential imaging strategies to multiplex. Here, we introduce blinkingbased multiplexing (BBM), a simple approach to differentiate spectrally overlapped emitters based solely on their intrinsic blinking dynamics. The blinking dynamics of hundreds of rhodamine 6G and CdSe/ZnS quantum dots on glass are obtained using the same acquisition settings and analyzed with a change point detection algorithm. Although substantial blinking heterogeneity is observed, the analysis yields a blinking metric with 93.5% classification accuracy. We further show that BBM with up to 96.6% accuracy is achieved by using a deep learning algorithm for classification. This proof-of-concept study demonstrates that a single emitter can be accurately classified based on its intrinsic blinking dynamics and without the need to probe its spectral color.
@article{desalvo2022blinking, title = {Blinking-based multiplexing: a new approach for differentiating spectrally overlapped emitters}, author = {DeSalvo, Grace A and Hoy, Grayson R and Kogan, Isabelle M and Li, John Z and Palmer, Elise T and Luz-Ricca, Emilio and de Gialluly, Paul Scemama and Wustholz, Kristin L}, journal = {The Journal of Physical Chemistry Letters}, volume = {13}, number = {22}, pages = {5056--5060}, year = {2022}, month = jun, publisher = {ACS Publications}, }